Isolation of Antibiotic Resistance organisms using gradient plate method
Aim
To isolate antibiotic resistant mutant organisms using gradient plate method
Principle:
Mutation is a heritable change in the nucleotide sequence of DNA. Mutations may be characterized according to either the kind of genotypic change that has occurred or their phenotypic consequences. Mutations can alter the phenotype of a microorganism in several different ways. Morphological mutations change the microorganism’s colonial or cellular morphology. Nutritional or biochemical variation may occur in a gene that encodes an enzyme involved in a metabolic pathway of amino acid synthesis. Changes in gene regulation occurs when mutation occur in a gene encoding a transcription factor. Lethal mutations prevent the reproducing capability of the organism, and when expressed, it results in the death of the microorganism.
Gradient Plate Technique:
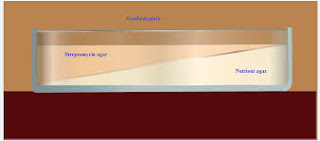
An excellent way to determine the ability of organisms to produce mutants that are resistant to antibiotic is to grow them on a gradient plate of a particular antibiotic. The gradient plate consists of two wedges like layers of media: a bottom layer of plain nutrient agar and top layer of antibiotic with nutrient agar. The antibiotic in the top layer, diffuse into the bottom layer producing a gradient of antibiotic concentration from low to high. A gradient plate is made by using Streptomycin in the medium. E. coli, which is normally sensitive to Streptomycin, will be spread over the surface of the plate and incubated for 24 to 72 hours. After incubation colonies will appear on both the gradients. The colonies develop in the high concentration are resistant to the action of Streptomycin, and are considered as Streptomycin resistant mutants. For isolation of antibiotic resistant of gram negative enteric bacteria, the antibiotics commonly used are Rifampicin, Streptomycin, and Erythromycin etc.
Materials required:
24 hour old nutrient broth culture of Escherichia coli.
Two nutrient agar deep tubes (10 ml per tube/culture).
1% Streptomycin sulphate solution (100 µg/ml).
A beaker with 90% ethanol.
Sterile Petri plates.
Sterile 1 ml pipette.
Glass rod spreader.
Water bath.
Procedure:
I) Preparation of gradient plate:
Melt two nutrient agar plates maintained at 96 degree C and cool to 55degree C
Pour the contents of one agar tube into a sterile petriplate. Allow the medium to solidify in a slanting position by placing either a glass rod under one side.
After the agar medium is solidified remove the glass rod and place the plate in the horizontal position.
Pipette out 0.1mL of 1% Streptomycin solution into the second tube of the second nutrient agar medium.
Rotate the tube between the palms and pour contents to cover the gradient layer agar and allow to the medium to solidify on a level table.
Label the low and high antibiotic concentration area on the bottom of the plate.
II) Inoculation of culture:
Pipette out 200µl (0.2ml) of the overnight Escherichia coli culture onto the gradient plate after 24 hours of its preparation.
Spread the inoculums evenly over the agar surface With a sterile bent glass rod by rotating the plate.
Incubate the inoculated plate in an inverted position at 37o C for 48-72 hours.
Observe the plate for appearance of E.coli colonies in the area of low streptomycin concentration (LSC) and high streptomycin concentration (HSC) and record the results.
Results:
Colonies which appear in the area of high concentration streptomycin region will be streptomycin resistant mutants.
III. Confirming presence of Streptomycin resistant colonies of E.coli.
Select and mark an isolated colony of E.coli in the HSC region of the Nutrient agar plate.
Pick the selected colony with a sterile inoculating loop and streak on to second gradient plate towards the HSC region.
Repeat this step with one or two colonies of streptomycin resistant mutants from the HSC region.
Incubate the inoculated plates in an inverted position at 37o C for 24-72 hours.
Observe the growth of streaked colonies towards the HSC region.
Results:
Growth of E.coli colonies in HSC area indicates the successful isolation of streptomycin resistant mutants.
Reference
- Brown E. Alfred, Benson’s Microbiological Applications, ninth edition, McGraw Hill Publication
- Prescott M. Lansing, Harley P. John, Klein A. Donald, Laboratory Exercises in Microbiology, fifth edition, McGraw-Hill college division.
- Aneja K R. Experiments in microbiology, plant pathology and biotechnology, fourth edition, New Age International (P) Limited .Publishers.
- Prescott M. Lansing, Harley P. John, Klein A. Donald, Microbiology, sixth edition, McGraw-Hill Higher Education
- Lederberg, J and Lederberg, EM (1952) Replica plating and indirect selection of bacterial mutants. J Bacteriol. 63: 399–406
- Madigan & Martinko, Brock Biology of Microorganisms, Eleventh Edition, Pearson Prentice Hall, Inc.